|
Size: 7789
Comment:
|
Size: 8357
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 76: | Line 76: |
| * {{{ncx_id}}} ncx by 1 vector of integers. | * {{{ncx_id1_id2}}} ncx by 2 matrix of integers. |
| Line 80: | Line 80: |
| * {{{ncn_id}}} ncn by 1 vector of integers. | * {{{ncn_id1_id2}}} ncn by 2 matrix of integers. |
| Line 92: | Line 92: |
| '''Remark 4.''' {{{xxx_id}}} vectors select subset of {{{xparam1}}} (for instance, {{{xparam_1(np_id)}}} selects all the lines of {{{xparam1}}} corresponding to the estimated "deep" parameters). | '''Remark 4.''' {{{np_id}}} vectors select subset of {{{xparam1}}} (for instance, {{{xparam_1(np_id)}}} selects all the lines of {{{xparam1}}} corresponding to the estimated "deep" parameters). |
| Line 94: | Line 94: |
| '''Remark 5.''' {{{nvx_id}}} and {{{nvn_id}}} vectors select subset of the diagonal elements of {{{Q}}} and, respectively, {{{H}}}. (for instance {{{Q(ncx_id,ncx_id)}}} selects the diagonal elements of {{{Q}}} corresponding to the estimated variance of shocks). '''Remark 6.''' {{{ncx_id1_id2}}} and {{{ncn_id1_id2}}} matrices select subset of the elements of {{{Q}}} and, respectively, {{{H}}}. (for instance {{{Q(ncx_id1_id2(1,1),ncx_id1_id2(1,2))}}} selects one element of {{{Q}}} corresponding to the first estimated covariance between two shocks). |
Proposed specification for estimation functions
This includes factorization of common code and extensions for (unexpected) structural change
Estimated parameters description
xparam1 is a column n by 1 vector holding the current values of the estimated parameters (n is the number of parameters).
parameters_description is a (global?) strucure of arrays describing the estimated parameters with the following fields:
type is a n by one vector of integers (1,2,3,4,5) where:
1 -> standard deviation of a structural shock.
2 -> standard deviation of a measurement error.
3 -> correlation betwwen two structural shocks.
4 -> correlation between two measurement errors.
5 -> "deep" parameter.
ID1 is a n by 1 vector of indices targeting to the (parameter, exogenous variable or endogenous variable) names (in M_.param_names, M_.exo_names and M_.endo_names).
ID2 is a n by 1 vector of NaNs or indices (second exogenous or endogenous variable index for the estimated correlations).
StartPeriod n by 1 (beginning of sub-period)
EndPeriod n by 1 (end of sub-period)
Prior is a sub-structure describing the prior belief on a parameter. The fiels are:
shape is an n by 1 vector of integers (1,2,3,4,5,6) where:
1 -> Beta density.
2 -> Gamma density.
3 -> Gaussian density.
4 -> Inverse gamma (type 1) density.
5 -> Uniform density.
6 -> Inverse gamma (type 2) density.
mean is an n by 1 vector of doubles (mean of the prior density).
mode is an n by 1 vector of doubles (mode of the prior density).
standard is an n by 1 vector of doubles (standard deviation of the prior density).
lower_bound is an n by 1 vector of doubles (and Inf/-Inf) (prior density domain).
upper_bound is an n by 1 vector of doubles (and Inf/-Inf) (prior density domain).
fhp is an n by 1 vector of doubles (first hypermarameter
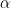 for the beta and gamma priors,
for the beta and gamma priors,  for the inverse gamma prior, lower bound for the uniform prior).
for the inverse gamma prior, lower bound for the uniform prior). shp is an n by 1 vector of doubles (second hypermarameter
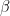 for the beta and gamma priors,
for the beta and gamma priors,  for the inverse gamma prior, upper bound for the uniform prior).
for the inverse gamma prior, upper bound for the uniform prior).
lower_bound n ny 1 vector of doubles (effective lower bound for the estimated parameter).
upper_bound n by 1 vector of doubles (effective upper bound for the estimated parameter).
nvx integer scalar (number of estimated structural shock standard deviations).
nvx_id nvx by 1 vector of integers.
nvn integer scalar (number of estimated measurement error standard deviations).
nvn_id nvn by 1 vector of integers.
ncx integer scalar (number of estimated structural shock correlations).
ncx_id1_id2 ncx by 2 matrix of integers.
ncn integer scalar (number of estimated measurement error correlations).
ncn_id1_id2 ncn by 2 matrix of integers.
np integer scalar (number of estimated "deep" parameters).
np_id np by 1 vector of integers.
Remark 1. The considered subsamples for different parameters may be different.
Remark 2. StartPeriod and EndPeriod specify the closed sub sample used to estimate the parameter. If these two fields are empty the parameter is estimated using the whole sample.
Remark 3. The field Prior is empty if the model is estimated by maximum lilkelihood, simulated moments or indirect inference.
Remark 4. np_id vectors select subset of xparam1 (for instance, xparam_1(np_id) selects all the lines of xparam1 corresponding to the estimated "deep" parameters).
Remark 5. nvx_id and nvn_id vectors select subset of the diagonal elements of Q and, respectively, H. (for instance Q(ncx_id,ncx_id) selects the diagonal elements of Q corresponding to the estimated variance of shocks).
Remark 6. ncx_id1_id2 and ncn_id1_id2 matrices select subset of the elements of Q and, respectively, H. (for instance Q(ncx_id1_id2(1,1),ncx_id1_id2(1,2)) selects one element of Q corresponding to the first estimated covariance between two shocks).
Functions
- Likelihood/posterior evaluation (separate two functions ?)
- INPUTS: xparam1, data, parameters_specification, ys, coeff_trends, M_.params, M_.Sigma_e, M_.H, list_of_observed_variables
- OUTPUTS: likelihood/posterior value, info
- Parameter updates (for period 1):
- INPUTS: xparam1, ys, coeff_trends,M_.params, M_.Sigma_e, M_.H
- OUTUTS: coeff_trends, params, Sigma_e, H
- reduced form (for period 1):
- INPUTS: params
- OUTPUTS: T R ys
- Computes steady state
- Computes linear solution for state variables + observed variables
- data_filtering (for period 1):
- INPUTS: data, ys, coeff_trends, StartPeriod, EndPeriod
- OUTPUTS: centered_data for subperiod
- Kalman filter initialization
- INPUTS: Sigma_e T R list_of_observed_variables
- OUTPUTS: a0 P0 Z
- Remark: Z is computed only for nonstationary models. mfs is computed once before these functions
- Diffuse Kalman filter recursions takes place here ?
- Kalman filter
- INPUTS: a0 Po Z T R centered_data Sigma_e H - OUTPUTS: log-likelihood, a(t|t-1), P(t|t-1)
if period < NP:
- parameter updates
- reduced form
- data_filtering
- priors evaluation
Additional Description Notes
Parameters and Sub-Samples
The initialization of estimation must detect the set of sub-samples where all parameters remain constant (such that max(StartPeriod)< t < min(EndPeriod) for the sub-sample) and make a table of which parameter is active in which subsample. For this, we need an additional component to the structure describing the parameters.
There is a prior for each sub-sample where the parameter remains constant. These sub-samples can be the union of several elementary sub-sampels defined as above.
Kalman
As change in parameters is not expected by the agents, the solution is very simple, we simply change the values of the system matrices in the Kalman filter within each sub-sample.
However, it would be too complicated to write a Kalman filter routine with changing matrices, so we prefer repeat the basic steps of the current implementation of DsgeLikelihood for each sub-sample during which all parameters remain unchanged.
The consistency of the transition from one sub-period to the next is insured by the proper initialization of initial state (a0) and initial covariance matrix (P0), that are taken from the last value of the previous sub-sample.
Likelihood Calculation
The likelihood (posterior) for the entire sample is simply computed as the sum of the likelihood for each sub-sample (weighted for the length of the sub-sample?).
From an estimation point of view, the changing parameters are treated as different parameters, so in the prior, constant parameters count for one, and each different values of a parameter count each for one as well.
Therefore, the value of log prior is not computed in the loop but only once at the end of DsgeLikelihood for all parameters.
Because we compute the likelihood (posterior) for the entire sample, introducing changing parameters doesn't modify the way the optimizers or the MCMC procedures calls DsgeLikelihood.