|
Size: 7478
Comment:
|
Size: 7766
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| ## page was renamed from FastDeterministicSimulation | |
| Line 2: | Line 3: |
| This page documents the ~+{{{model}}} +~and ~+{{{simul}}}+~ options used to speed-up deterministic simulation in middle and large scale models. Two groups of options are considered: | |
| Line 3: | Line 5: |
| This page documents the ~+ {{{model}}} +~ and ~+ {{{simul}}}+~ options used to speed-up deterministic simulation in middle and large scale models. Two groups of options are considered: | |
| Line 6: | Line 7: |
| Line 8: | Line 8: |
| Line 10: | Line 9: |
| Line 18: | Line 16: |
| === sparse_dll === Dynare performs also a block-decomposition of the model and the preprocessor creates two C-files or a binary file containing the model code according to the "Compiler option" containing the code of the deterministic simulation. |
|
| Line 19: | Line 19: |
| === sparse_dll === Dynare performs also a block-decomposition of the model and the preprocessor creates two C-files or a binary file containing the model code according to the "Compiler option" containing the code of the deterministic simulation. |
|
| Line 27: | Line 24: |
| Line 31: | Line 27: |
| These options have to be used in conjunction with sparse_dll option. Two values could be used ~+{{{GCC_compiler}}}+~ or ~+{{{NO_compiler}}}+~ (default value). | |
| Line 32: | Line 29: |
| These options have to be used in conjunction with sparse_dll option. Two values could be used ~+{{{GCC_compiler}}}+~ and ~+{{{[NO_compiler]}}}+~ (default value). * ~+{{{GCC_compiler}}}+~: with the first option a C file the preprocessor creates a C file containing the dynamic model which is compiled with the GCC compiler. * ~+{{{[NO_compiler]}}}+~: a binary file containing the code of the dynamic model is created. During the simulation step binary code is evaluated by ~+{{{simulate.dll}}}+~. |
* ~+{{{GCC_compiler}}}+~: with the first option the preprocessor creates a C file containing the dynamic model which is compiled with the GCC compiler. * ~+{{{NO_compiler}}}+~: a binary file containing the code of the dynamic model is created. During the simulation step binary code is evaluated by ~+{{{simulate.dll}}}+~. |
| Line 40: | Line 36: |
/!\ if you use ~+{{{GCC_compiler}}}+~ Gnumex has to be installed on your computer. |
/!\ if you use ~+{{{GCC_compiler}}}+~ Gnumex has to be installed on your computer. |
| Line 45: | Line 39: |
| Line 55: | Line 48: |
/!\ Note that the jacobian is evaluated considering the initial value of the parameters provided in the mod file. If the parameters value are modified (the model is estimated for example) before the ~+{{{simul}}}+~ command, some components of the jacobian could be wrongly discarded. To prevent such a result you have to set the cutoff to O. |
/!\ Note that the jacobian is evaluated considering the initial value of the parameters provided in the mod file. If the parameters value are modified (the model is estimated for example) before the ~+{{{simul}}}+~ command, some components of the jacobian could be wrongly discarded. To prevent such a result you have to set the cutoff to 0. |
| Line 61: | Line 51: |
| At each iteration of the deterministic simulation, if the linear approximation of the model is solve using a sparse gaussian elimination, two criterion could be used to select the variable to substitute: | |
| Line 62: | Line 53: |
| At each iteration of the deterministic simulation, if the linear approximation of the model is solve using a sparse Gaussian elimination, two criterion could be used to select the variable to substitute : * The maximum pivot to get the most accurate solution (quite standard in Gaussian Elimination or LU decomposition). * The Markowitz criterion, to prevent an excessive sparsity reduction. The number of new non-zero elements created during the variable substitution is related to the number of non zero elements in its row. In this case we select the variable associated to the minimum non-zero elements in its row. Markowitz option |
* The maximum pivot to get the most accurate solution (quite standard in Gaussian Elimination or LU decomposition). * The Markowitz criterion, to prevent an excessive sparsity reduction. The number of new non-zero elements created during the variable substitution is related to the number of non zero elements in its row. In this case we select the variable associated to the minimum non-zero elements in its row. Markowitz option |
| Line 68: | Line 58: |
| {{{#!latex | {{{ #!latex |
| Line 73: | Line 65: |
| With: | |
| Line 74: | Line 67: |
| With: * [[latex($\lambda_j$)]]: the value of jth pivot; * [[latex($Sup_{i} \left | \lambda_i\right |$)]]: the highest pivot among the N remaining pivots ; * [[latex($n_j$)]]: the number of non zeros elements in the row related to the jth pivot ; * [[latex($n_j$)]]: the highest number of non zeros elements in the row related to remaining pivots ; * [[latex($\mu$)]] : the value of the Markowitz option. As [[latex($\mu \rightarrow \infty $)]] the Markowitz criteria prevails. At the opposite if [[latex($\mu \rightarrow 0 $)]] the usual maximum pivot criteria prevails. |
* <<latex($\lambda_j$)>>: the value of jth pivot; * <<latex($Sup_{i} \left | \lambda_i\right |$)>>: the highest absolute value of the pivot among the N remaining pivots ; * <<latex($n_j$)>>: the number of non zeros elements in the row related to the jth pivot ; * <<latex($n_j$)>>: the highest number of non zeros elements in the row related to remaining pivots ; * <<latex($\mu$)>> : the value of the Markowitz option. As <<latex($\mu \rightarrow \infty $)>> the Markowitz criteria prevails. At the opposite if <<latex($\mu \rightarrow 0 $)>> the usual maximum pivot criteria prevails. |
| Line 90: | Line 81: |
/!\This option is only used in conjunction with sparse_dll option. |
/!\ This option is only used in conjunction with sparse_dll option. |
| Line 94: | Line 84: |
| The ~+{{{model_info}}} +~command provides information about | |
| Line 95: | Line 86: |
| The ~+{{{model_info}}}+~ command provides information about * the normalization of the model: an endogenous variable is attributed to each equation of the model; * the block structure of the model: for each block {model_info} indicates its type, the equations number and endogenous variables belonging to this block. |
* the normalization of the model: an endogenous variable is attributed to each equation of the model; * the block structure of the model: for each block ~+{{{model_info}}} +~indicates its type, the equations number and endogenous variables belonging to this block. There are five different types of blocks depending on the simulation method used: |
| Line 99: | Line 90: |
| There are five different types of blocks depending on the simulation method used: * EVALUATE FORWARD: in this case the block contains only equations where endogenous variable attributed to the equation appears currently on the left hand side and where no forward looking endogenous variables appear. [[latex($y_{j,t} = f_j \left (y_t, y_{t-1}, …, y_{t-k} \right) $)]]. * EVALUATE BACKWARD: the block contains only equations where endogenous variable attributed to the equation appears currently on the left hand side and where no backward looking endogenous variables appear. [[latex($y_{j,t} = f_j \left (y_t, y_{t+1}, …, y_{t+k} \right) $)]]. * SOLVE FORWARD x: the block contains only equations where endogenous variable attributed to the equation does not appear currently on the left hand side and where no forward looking endogenous variables appear. [[latex($ g_j \left (y_{j,t}, y_t, y_{t-1}, …, y_{t-k} \right) =0 $)]]. x is equal to SIMPLE if the block has only one equation. If several equation appears in the block x is equal to COMPLETE. * SOLVE FORWARD x: the block contains only equations where endogenous variable attributed to the equation does not appear currently on the left hand side and where no backward looking endogenous variables appear. [[latex($ g_j \left (y_{j,t}, y_t, y_{t+1}, …, y_{t+k} \right) =0 $)]]. x is equal to SIMPLE if the block has only one equation. If several equation appears in the block x is equal to COMPLETE. * SOVE TWO BOUNDARIES x: the block contains equations depending on both forward and backward variables. [[latex($ g_j \left (y_{j,t}, y_t, y_{t-1}, …, y_{t-k} \ ,y_t, y_{t+1}, …, y_{t+k} \right) =0 $)]]. x is equal to SIMPLE if the block has only one equation. If several equation appears in the block x is equal to COMPLETE. |
* EVALUATE FORWARD: in this case the block contains only equations where endogenous variable attributed to the equation appears currently on the left hand side and where no forward looking endogenous variables appear. <<latex($y_{j,t} = f_j \left (y_t, y_{t-1}, …, y_{t-k} \right) $)>>. * EVALUATE BACKWARD: the block contains only equations where endogenous variable attributed to the equation appears currently on the left hand side and where no backward looking endogenous variables appear. <<latex($y_{j,t} = f_j \left (y_t, y_{t+1}, …, y_{t+k} \right) $)>>. * SOLVE FORWARD x: the block contains only equations where endogenous variable attributed to the equation does not appear currently on the left hand side and where no forward looking endogenous variables appear. <<latex($ g_j \left (y_{j,t}, y_t, y_{t-1}, …, y_{t-k} \right) =0 $)>>. x is equal to SIMPLE if the block has only one equation. If several equation appears in the block x is equal to COMPLETE. * SOLVE FORWARD x: the block contains only equations where endogenous variable attributed to the equation does not appear currently on the left hand side and where no backward looking endogenous variables appear. <<latex($ g_j \left (y_{j,t}, y_t, y_{t+1}, …, y_{t+k} \right) =0 $)>>. x is equal to SIMPLE if the block has only one equation. If several equation appears in the block x is equal to COMPLETE. * SOVE TWO BOUNDARIES x: the block contains equations depending on both forward and backward variables. <<latex($ g_j \left (y_{j,t}, y_t, y_{t-1}, …, y_{t-k} \ ,y_t, y_{t+1}, …, y_{t+k} \right) =0 $)>>. x is equal to SIMPLE if the block has only one equation. If several equation appears in the block x is equal to COMPLETE. |
| Line 118: | Line 102: |
| == simulate options == === method === This option indicates which linear solver will be implemented at each step of the Newton simulation of the stacked systems formed by the simultaneous blocks containing leads and lags on the endogenous variables. There are three values for this option if the ~+{{{sparse}}}+~ option is used in model command: |
|
| Line 119: | Line 106: |
| * LU: for the sparse LU decomposition (the default option); * GMRES: for the Generalized Minimal Residual method; * BICGSTAB: for the Stabilized Bi-Conjugate Gradient method. When sparse_dll option is used only SGE (for Sparse Gaussian Elimination) is available. It is the unique and default value of method in this case. |
|
| Line 120: | Line 111: |
| == simulate options == | === datafile === |
| Line 122: | Line 113: |
| === method === | If the variables of the model are not constant over time, their initial values, stored in a text file, could be loaded, using the datafile option, as initial values before a deteministic simulation. |
| Line 124: | Line 115: |
| This option indicates which linear solver will be implemented at each step of the Newton simulation of the stacked systems formed by the simultaneous blocks containing leads and lags on the endogenous variables. There are three values for this option if the ~+{{{sparse}}}+~ option is used in model command: * LU: for the sparse LU decomposition (the default option); * GMRES: for the Generalized Minimal Residual method; * BICGSTAB: for the Stabilized Bi-Conjugate Gradient method. |
{{{ model; ... end; |
| Line 129: | Line 120: |
| When sparse_dll option is used in model only SGE (for Sparse Gaussian Elimination is available). It is the unique and default value of method in this case. | simul(periods=80,datafile=mark3); }}} |
Fast Deterministic Simulation
This page documents the model and simul options used to speed-up deterministic simulation in middle and large scale models. Two groups of options are considered:
- The options and command taking advantage of the sparsity of the model: block decomposition and fill-in;
- The options indicating which algorithm is used to solve the model.
sparse model options and model_info command
sparse
Dynare performs a block-decomposition of the model and the preprocessor creates two m-files: ModFileName_static.m and ModFileName_dynamic.m containing the code for deterministic and stochastic simulation.
model(sparse); ... end;
sparse_dll
Dynare performs also a block-decomposition of the model and the preprocessor creates two C-files or a binary file containing the model code according to the "Compiler option" containing the code of the deterministic simulation.
model(sparse_dll); ... end;
![]() in the current version of Dynare this option is not compatible with the check command.
in the current version of Dynare this option is not compatible with the check command.
Compiler options
These options have to be used in conjunction with sparse_dll option. Two values could be used GCC_compiler or NO_compiler (default value).
GCC_compiler: with the first option the preprocessor creates a C file containing the dynamic model which is compiled with the GCC compiler.
NO_compiler: a binary file containing the code of the dynamic model is created. During the simulation step binary code is evaluated by simulate.dll.
model(sparse_dll,GCC_compiler); ... end;
![]() if you use GCC_compiler Gnumex has to be installed on your computer.
if you use GCC_compiler Gnumex has to be installed on your computer.
cutoff
To get ride of nearly zero elements in the jacobian a threshold is used to determine whether a jacobian element has to be considered as null or not. If the absolute value of a jacobian element is below the threshold then it is considered as null and is discarded form the sparse representation of the jacobian. Eliminating nearly zero elements improves the sparsity of the model and hence the simulation time. Note that if the cutoff value is too high then the model properties could be strongly modified and the simulation could failed.
The default value of the cutoff is 1e-15.
model(sparse,cutoff=1e-13); ... end;
![]() Note that the jacobian is evaluated considering the initial value of the parameters provided in the mod file. If the parameters value are modified (the model is estimated for example) before the simul command, some components of the jacobian could be wrongly discarded. To prevent such a result you have to set the cutoff to 0.
Note that the jacobian is evaluated considering the initial value of the parameters provided in the mod file. If the parameters value are modified (the model is estimated for example) before the simul command, some components of the jacobian could be wrongly discarded. To prevent such a result you have to set the cutoff to 0.
markowitz
At each iteration of the deterministic simulation, if the linear approximation of the model is solve using a sparse gaussian elimination, two criterion could be used to select the variable to substitute:
- The maximum pivot to get the most accurate solution (quite standard in Gaussian Elimination or LU decomposition).
- The Markowitz criterion, to prevent an excessive sparsity reduction. The number of new non-zero elements created during the variable substitution is related to the number of non zero elements in its row. In this case we select the variable associated to the minimum non-zero elements in its row.
Markowitz option
The pivot is selected according to the highest value among j=1,…,N (where N is the number of possible pivots) of the following criteria:
![\[
\frac{\frac{\left | \lambda_j\right |}{Sup_{i} \left | \lambda_i\right |} }{\left ( \frac{ n_j }{ Sup_{i} n_i} \right )^\mu}
\] \[
\frac{\frac{\left | \lambda_j\right |}{Sup_{i} \left | \lambda_i\right |} }{\left ( \frac{ n_j }{ Sup_{i} n_i} \right )^\mu}
\]](/DynareWiki/FastDeterministicSimulationAndSteadyStateComputation?action=AttachFile&do=get&target=latex_7a35ec8a6b4af645da8e3f714cc1ec54ef9f92ce_p1.png)
With:
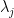 : the value of jth pivot;
: the value of jth pivot; 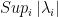 : the highest absolute value of the pivot among the N remaining pivots ;
: the highest absolute value of the pivot among the N remaining pivots ; 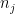 : the number of non zeros elements in the row related to the jth pivot ;
: the number of non zeros elements in the row related to the jth pivot ; 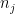 : the highest number of non zeros elements in the row related to remaining pivots ;
: the highest number of non zeros elements in the row related to remaining pivots ; 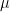 : the value of the Markowitz option.
: the value of the Markowitz option.
As 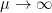 the Markowitz criteria prevails. At the opposite if
the Markowitz criteria prevails. At the opposite if 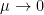 the usual maximum pivot criteria prevails.
the usual maximum pivot criteria prevails.
The default value is 0.5
model(sparse_dll,markowitz=2); ... end;
![]() This option is only used in conjunction with sparse_dll option.
This option is only used in conjunction with sparse_dll option.
model_info command
The model_info command provides information about
- the normalization of the model: an endogenous variable is attributed to each equation of the model;
the block structure of the model: for each block model_info indicates its type, the equations number and endogenous variables belonging to this block.
There are five different types of blocks depending on the simulation method used:
EVALUATE FORWARD: in this case the block contains only equations where endogenous variable attributed to the equation appears currently on the left hand side and where no forward looking endogenous variables appear.
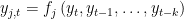 .
. EVALUATE BACKWARD: the block contains only equations where endogenous variable attributed to the equation appears currently on the left hand side and where no backward looking endogenous variables appear.
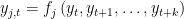 .
. SOLVE FORWARD x: the block contains only equations where endogenous variable attributed to the equation does not appear currently on the left hand side and where no forward looking endogenous variables appear.
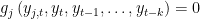 . x is equal to SIMPLE if the block has only one equation. If several equation appears in the block x is equal to COMPLETE.
. x is equal to SIMPLE if the block has only one equation. If several equation appears in the block x is equal to COMPLETE. SOLVE FORWARD x: the block contains only equations where endogenous variable attributed to the equation does not appear currently on the left hand side and where no backward looking endogenous variables appear.
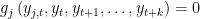 . x is equal to SIMPLE if the block has only one equation. If several equation appears in the block x is equal to COMPLETE.
. x is equal to SIMPLE if the block has only one equation. If several equation appears in the block x is equal to COMPLETE. SOVE TWO BOUNDARIES x: the block contains equations depending on both forward and backward variables.
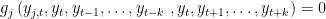 . x is equal to SIMPLE if the block has only one equation. If several equation appears in the block x is equal to COMPLETE.
. x is equal to SIMPLE if the block has only one equation. If several equation appears in the block x is equal to COMPLETE.
model(sparse); ... end; model_info;
simulate options
method
This option indicates which linear solver will be implemented at each step of the Newton simulation of the stacked systems formed by the simultaneous blocks containing leads and lags on the endogenous variables. There are three values for this option if the sparse option is used in model command:
- LU: for the sparse LU decomposition (the default option);
- GMRES: for the Generalized Minimal Residual method;
- BICGSTAB: for the Stabilized Bi-Conjugate Gradient method.
When sparse_dll option is used only SGE (for Sparse Gaussian Elimination) is available. It is the unique and default value of method in this case.
datafile
If the variables of the model are not constant over time, their initial values, stored in a text file, could be loaded, using the datafile option, as initial values before a deteministic simulation.
model; ... end; simul(periods=80,datafile=mark3);