Description
The equations of the model are written in a block delimited by
model and end keywords.
There must be as many equations as there are endogenous variables in the
model, except when computing the unconstrained optimal policy with
ramsey_model, ramsey_policy or discretionary_policy.
The syntax of equations must follow the conventions for MODEL_EXPRESSION as described in Expressions. Each equation must be terminated by a semicolon (‘;’). A normal equation looks like:
MODEL_EXPRESSION = MODEL_EXPRESSION; |
When the equations are written in homogenous form, it is possible to omit the ‘=0’ part and write only the left hand side of the equation. A homogenous equation looks like:
MODEL_EXPRESSION; |
Inside the model block, Dynare allows the creation of model-local
variables, which constitute a simple way to share a common expression
between several equations. The syntax consists of a pound sign
(#) followed by the name of the new model local variable (which
must not be declared as in Variable declarations), an equal
sign, and the expression for which this new variable will stand. Later
on, every time this variable appears in the model, Dynare will
substitute it by the expression assigned to the variable. Note that the
scope of this variable is restricted to the model block; it cannot be
used outside. A model local variable declaration looks like:
# VARIABLE_NAME = MODEL_EXPRESSION; |
It is possible to tag equations written in the model block. A tag can serve
different purposes by allowing the user to attach arbitrary informations to each
equation and to recover them at runtime. For instance, it is possible to name the
equations with a name-tag, using a syntax like:
mode;
...
[name = 'Budget constraint']
c + k = k^theta*A;
...
end;
|
Here, name is the keyword indicating that the tag names the equation. If an equation
of the model is tagged with a name, the resid command
will display the name of the equations (which may be more informative than the
equation numbers) in addition to the equation number. Several tags for one equation can be separated using a comma.
mode;
...
[name='Taylor rule',mcp = 'r > -1.94478']
r = rho*r(-1) + (1-rho)*(gpi*Infl+gy*YGap) + e;
...
end;
|
More information on tags is available on the DynareWiki wiki.
Options
-
linear Declares the model as being linear. It spares oneself from having to declare initial values for computing the steady state of a stationary linear model. This option can’t be used with non-linear models, it will NOT trigger linearization of the model.
-
use_dll Instructs the preprocessor to create dynamic loadable libraries (DLL) containing the model equations and derivatives, instead of writing those in M-files. You need a working compilation environment, i.e. a working
mexcommand (see Compiler installation for more details). On MATLAB for Windows, you will need to also pass the compiler name at the command line. Using this option can result in faster simulations or estimations, at the expense of some initial compilation time.(2)-
block Perform the block decomposition of the model, and exploit it in computations (steady-state, deterministic simulation, stochastic simulation with first order approximation and estimation). See Dynare wiki for details on the algorithms used in deterministic simulation and steady-state computation.
-
bytecode Instead of M-files, use a bytecode representation of the model, i.e. a binary file containing a compact representation of all the equations.
-
cutoff = DOUBLE Threshold under which a jacobian element is considered as null during the model normalization. Only available with option
block. Default:1e-15-
mfs = INTEGER Controls the handling of minimum feedback set of endogenous variables. Only available with option
block. Possible values:-
0 All the endogenous variables are considered as feedback variables (Default).
-
1 The endogenous variables assigned to equation naturally normalized (i.e. of the form
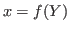 where
where 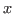 does not appear in
does not appear in
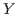 ) are potentially recursive variables. All the other variables
are forced to belong to the set of feedback variables.
) are potentially recursive variables. All the other variables
are forced to belong to the set of feedback variables.
-
2 In addition of variables with
mfs = 1the endogenous variables related to linear equations which could be normalized are potential recursive variables. All the other variables are forced to belong to the set of feedback variables.-
3 In addition of variables with
mfs = 2the endogenous variables related to non-linear equations which could be normalized are potential recursive variables. All the other variables are forced to belong to the set of feedback variables.
-
-
no_static Don’t create the static model file. This can be useful for models which don’t have a steady state.
-
differentiate_forward_vars -
differentiate_forward_vars = ( VARIABLE_NAME [VARIABLE_NAME …] ) Tells Dynare to create a new auxiliary variable for each endogenous variable that appears with a lead, such that the new variable is the time differentiate of the original one. More precisely, if the model contains
x(+1), then a variableAUX_DIFF_VARwill be created such thatAUX_DIFF_VAR=x-x(-1), andx(+1)will be replaced withx+AUX_DIFF_VAR(+1).The transformation is applied to all endogenous variables with a lead if the option is given without a list of variables. If there is a list, the transformation is restricted to endogenous with a lead that also appear in the list.
This option can useful for some deterministic simulations where convergence is hard to obtain. Bad values for terminal conditions in the case of very persistent dynamics or permanent shocks can hinder correct solutions or any convergence. The new differentiated variables have obvious zero terminal conditions (if the terminal condition is a steady state) and this in many cases helps convergence of simulations.
-
parallel_local_files = ( FILENAME [, FILENAME]… ) Declares a list of extra files that should be transferred to slave nodes when doing a parallel computation (see section Parallel Configuration).
Example 1: elementary RBC model
var c k; varexo x; parameters aa alph bet delt gam; model; c = - k + aa*x*k(-1)^alph + (1-delt)*k(-1); c^(-gam) = (aa*alph*x(+1)*k^(alph-1) + 1 - delt)*c(+1)^(-gam)/(1+bet); end; |
Example 2: use of model local variables
The following program:
model; # gamma = 1 - 1/sigma; u1 = c1^gamma/gamma; u2 = c2^gamma/gamma; end; |
…is formally equivalent to:
model; u1 = c1^(1-1/sigma)/(1-1/sigma); u2 = c2^(1-1/sigma)/(1-1/sigma); end; |
Example 3: a linear model
model(linear); x = a*x(-1)+b*y(+1)+e_x; y = d*y(-1)+e_y; end; |