Description
Triggers the creation of an initialization file for, and the estimation
of, a Markov-switching SBVAR model. At the end of the run, the
![]() ,
, ![]() ,
, ![]() and
and ![]() matrices are contained
in the
matrices are contained
in the oo_.ms structure.
Options
General Options
-
file_tag = FILENAME The portion of the filename associated with this run. This will create the model initialization file,
init_<file_tag>.dat. Default:<mod_file>-
output_file_tag = FILENAME The portion of the output filename that will be assigned to this run. This will create, among other files,
est_final_<output_file_tag>.out,est_intermediate_<output_file_tag>.out. Default:<file_tag>-
no_create_init Do not create an initialization file for the model. Passing this option will cause the Initialization Options to be ignored. Further, the model will be generated from the output files associated with the previous estimation run (i.e.
est_final_<file_tag>.out,est_intermediate_<file_tag>.outorinit_<file_tag>.dat, searched for in sequential order). This functionality can be useful for continuing a previous estimation run to ensure convergence was reached or for reusing an initialization file. NB: If this option is not passed, the files from the previous estimation run will be overwritten. Default:off(i.e. create initialization file)
Initialization Options
-
coefficients_prior_hyperparameters = [DOUBLE1 DOUBLE2 DOUBLE3 DOUBLE4 DOUBLE5 DOUBLE6] Sets the hyper parameters for the model. The six elements of the argument vector have the following interpretations:
-
Position Interpretation-
1 Overall tightness for
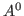 and
and 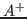
-
2 Relative tightness for
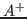
-
3 Relative tightness for the constant term
-
4 Tightness on lag decay (range: 1.2 - 1.5); a faster decay produces better inflation process
-
5 Weight on nvar sums of coeffs dummy observations (unit roots)
-
6 Weight on single dummy initial observation including constant
Default:
[1.0 1.0 0.1 1.2 1.0 1.0]-
-
freq = INTEGER |monthly|quarterly|yearly Frequency of the data (e.g.
monthly,12). Default:4-
initial_year = INTEGER The first year of data. Default:
none-
initial_subperiod = INTEGER The first period of data (i.e. for quarterly data, an integer in [
1,4]). Default:1-
final_year = INTEGER The last year of data. Default: Set to encompass entire dataset.
-
final_subperiod = INTEGER The final period of data (i.e. for monthly data, an integer in [
1,12]. Default: When final_year is also missing, set to encompass entire dataset; when final_year is indicated, set to the maximum number of subperiods given the frequency (i.e. 4 for quarterly data, 12 for monthly,...).-
datafile = FILENAME See datafile.
-
xls_sheet = NAME See xls_sheet.
-
xls_range = RANGE See xls_range.
-
nlags = INTEGER The number of lags in the model. Default:
1-
cross_restrictions Use cross
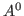 and
and 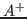 restrictions. Default:
restrictions. Default: off-
contemp_reduced_form Use contemporaneous recursive reduced form. Default:
off-
no_bayesian_prior Do not use Bayesian prior. Default:
off(i.e. use Bayesian prior)-
alpha = INTEGER Alpha value for squared time-varying structural shock lambda. Default:
1-
beta = INTEGER Beta value for squared time-varying structural shock lambda. Default:
1-
gsig2_lmdm = INTEGER The variance for each independent
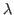 parameter under
parameter under
SimsZharestrictions. Default:50^2-
specification =sims_zha|none This controls how restrictions are imposed to reduce the number of parameters. Default:
Random Walk
Estimation Options
-
convergence_starting_value = DOUBLE This is the tolerance criterion for convergence and refers to changes in the objective function value. It should be rather loose since it will gradually be tightened during estimation. Default:
1e-3-
convergence_ending_value = DOUBLE The convergence criterion ending value. Values much smaller than square root machine epsilon are probably overkill. Default:
1e-6-
convergence_increment_value = DOUBLE Determines how quickly the convergence criterion moves from the starting value to the ending value. Default:
0.1-
max_iterations_starting_value = INTEGER This is the maximum number of iterations allowed in the hill-climbing optimization routine and should be rather small since it will gradually be increased during estimation. Default:
50-
max_iterations_increment_value = DOUBLE Determines how quickly the maximum number of iterations is increased. Default:
2-
max_block_iterations = INTEGER The parameters are divided into blocks and optimization proceeds over each block. After a set of blockwise optimizations are performed, the convergence criterion is checked and the blockwise optimizations are repeated if the criterion is violated. This controls the maximum number of times the blockwise optimization can be performed. Note that after the blockwise optimizations have converged, a single optimization over all the parameters is performed before updating the convergence value and maximum number of iterations. Default:
100-
max_repeated_optimization_runs = INTEGER The entire process described by max_block_iterations is repeated until improvement has stopped. This is the maximum number of times the process is allowed to repeat. Set this to
0to not allow repetitions. Default:10-
function_convergence_criterion = DOUBLE The convergence criterion for the objective function when
max_repeated_optimizations_runsis positive. Default:0.1-
parameter_convergence_criterion = DOUBLE The convergence criterion for parameter values when
max_repeated_optimizations_runsis positive. Default:0.1-
number_of_large_perturbations = INTEGER The entire process described by max_block_iterations is repeated with random starting values drawn from the posterior. This specifies the number of random starting values used. Set this to
0to not use random starting values. A larger number should be specified to ensure that the entire parameter space has been covered. Default:5-
number_of_small_perturbations = INTEGER The number of small perturbations to make after the large perturbations have stopped improving. Setting this number much above
10is probably overkill. Default:5-
number_of_posterior_draws_after_perturbation = INTEGER The number of consecutive posterior draws to make when producing a small perturbation. Because the posterior draws are serially correlated, a small number will result in a small perturbation. Default:
1-
max_number_of_stages = INTEGER The small and large perturbation are repeated until improvement has stopped. This specifics the maximum number of stages allowed. Default:
20-
random_function_convergence_criterion = DOUBLE The convergence criterion for the objective function when
number_of_large_perturbationsis positive. Default:0.1-
random_parameter_convergence_criterion = DOUBLE The convergence criterion for parameter values when
number_of_large_perturbationsis positive. Default:0.1